The Nuclefy Genomic DNA Purification Kit is designed for rapid and reliable sample lysis and purification of intact genomic DNA (gDNA) from a variety of biological sample types.
The protocol utilizes specifically formulated buffer systems allowing to process blood, tissues, and cultured cells. Supplemental protocols allow to process clinically-relevant samples such as saliva and buccal swabs, as well as bacteria, yeast and insects. In addition, it can also be used for classic gDNA cleanup.
Performance and Downstream Applications
The Nuclefy Genomic DNA Purification Kit generates excellent input material regarding DNA integrity and purity for downstream applications such as quantitative PCR and NGS library preparation.
DNA Integrity
Genomic DNA samples were analyzed regarding DNA integrity by loading 50 ng on a 1 % agarose gel. Genomic DNA purification was performed following the appropriate purification protocols. Three representative DNA obtained from tissue (chicken heart, lung and muscle), human whole blood, cattle blood, HeLa and NIH/3T3 cells, E.coli, R. terrae, saliva as well as buccal swab are shown.
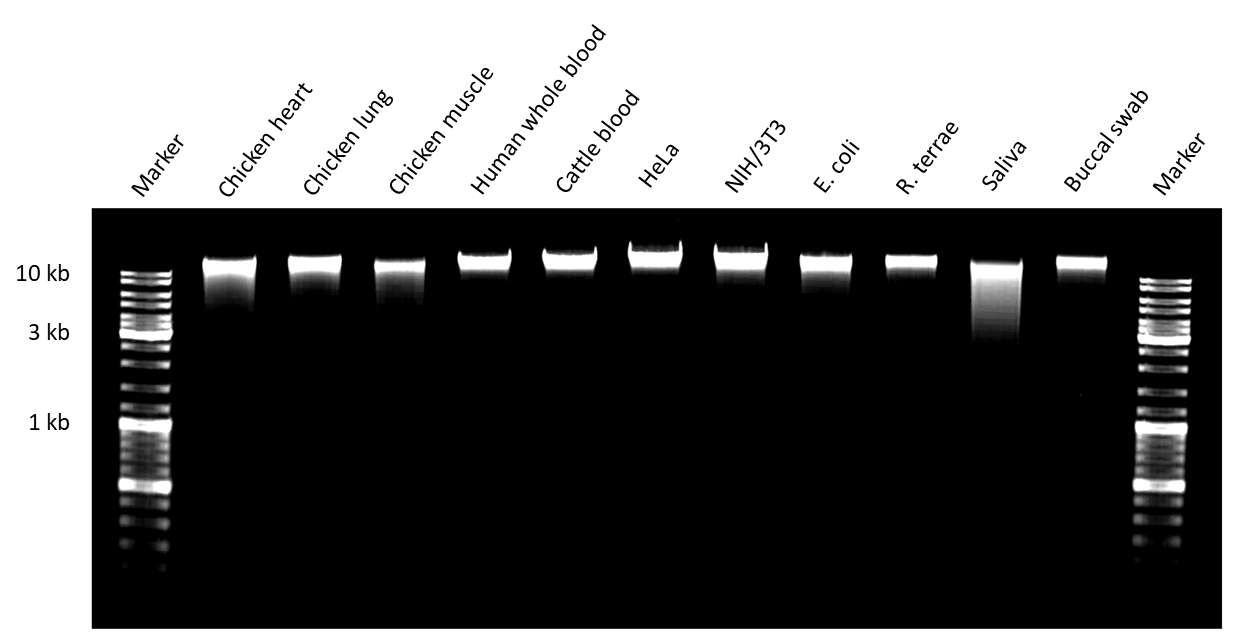
Figure 1. Integrity of gDNA isolated from representative sample types.
Quantitative PCR
Purified gDNA from several representative sample types was diluted to produce a four log range of input template concentrations. Results were obtained using primers targeting poultry- (chicken muscle), human- (whole blood, buccal swab, HeLa cells) as well as bacteria-specific (E. coli) genomic sequences for qPCR assays with the PhoenixDx® qPCR Mastermixes (Procomcure Biotech #PCCSKU12011) and cycled on Applied Biosystems™ QuantStudio 5 qPCR thermal cycler. Results confirm that eluted gDNA is highly pure and free from inhibitors, optimal for qPCR.
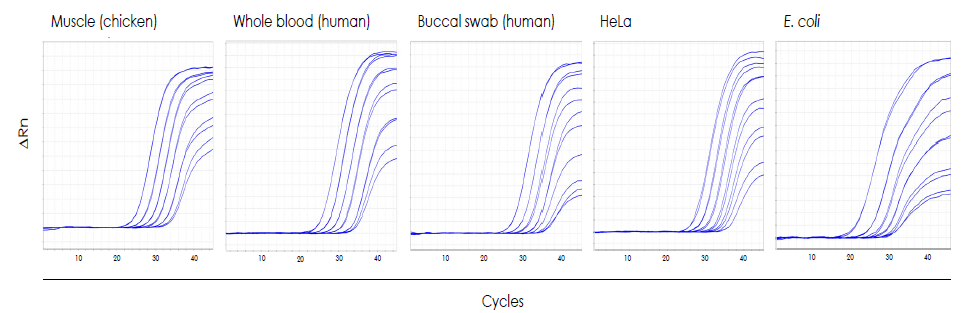
Figure 2. qPCR data using species-specific designed assays.
NGS Library Preparation
Genomic DNA was isolated from whole human blood with Nuclefy Genomic DNA Purification Kit (Procomcure Biotech #PCCSKU16073) and Monarch® Genomic DNA Purification Kit (NEB #T3010). Duplicate libraries were prepared from 200 ng human genomic DNA using the NEBNext Ultra II FS DNA Library Prep Kit for Illumina (NEB #E7805). Libraries were sequenced on an Illumina MiSeq®. Reads were mapped using BWA and GC coverage was calculated using Picard‘s CollectGCBiasMetrics (v2.26.2). Library yields of samples were assessed on a Caliper Labchip GX II using a High Sensitivity DNA Kit.

Figure 3. A GC bias plot. B Library yield.